
Mount Sinai Scientists Develops A New Reference Database (RepurposeDB) of Drug Repositioning and Decodes Molecular Code of Therapeutic Reuse
By: Vijay Soni, Ph.D. | Founder & C.E.O Scipreneur
The first centralized open-access reference database of drug repositioning investigations and compiled data on drugs, disease indications, drug targets, disease comorbidities and various annotations
The first centralized open-access reference database of drug repositioning investigations and compiled data on drugs, disease indications, drug targets, disease comorbidities and various annotations
February 21, 2017
A drug discovery project takes 8-12 years and costs 8-15 billion dollars. It relies on various avenues of pre-clinical development, biomedical research, medicinal chemistry, clinical trials, and healthcare. Drug development project is expensive medical research projects that aim to bring a product from ideation to market. Developing a novel drug for every disease is a challenge due to the limited availability of resources. Pharmaceutical companies also prioritize drug development projects based on disease prevalence and forecasting. Various constraints in drug discovery due to cost, resources cannot reliably meet the growing patient needs to develop therapies for various diseases. In this context, drug repositioning – scientific method to evaluate existing drugs and target biology for new indication could pave a way to find effective therapies. One of the scientific rationales of drug repositioning suggests the overlapping role (pleiotropic effect) of biomolecules contributing to pathophysiology process of multiple diseases.
RepurposeDB could help to develop predictive models of drug repositioning and can have major influence in personalized drug repositioning, an emerging therapeutic area in data-driven, individualized medicine.
RepurposeDB could help to develop predictive models of drug repositioning and can have major influence in personalized drug repositioning, an emerging therapeutic area in data-driven, individualized medicine.
Drug repositioning studies are emerging in the recent years, however, hampered by the lack of a centralized database as well as a lack of reporting standards for drug repositioning investigations. A team of scientists led by Dr. Joel T Dudley (Institute of Next Generation Healthcare, Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, Mount Sinai Health System) has now developed a new reference database of drug repositioning studies – RepurposeDB. RepurposeDB, a drug discovery resource funded by National Institutes of Health grants currently has 253 drugs [chemical compounds (74.30%) and biotech drugs (25.29%)] and 1125 disease indications. The database also offers computational drug discovery tools including drug similarity searches. The investigators also present a new standard “Minimum Information about Drug Repositioning Investigations (MIADRI)” for reporting the drug repositioning investigation. The RepurposeDB team believes MIADRI and the open access data policies would encourage the scientific and clinical investigators from around the world to crowdsource the drug repositioning into a centralized database. A manuscript describing the RepurposeDB, MIADRI, and analyses of the data with Dr. Shameer Khader as the primary author is available in the official journal of International Society for Computational Biology, Briefings in Bioinformatics – published by Oxford University Press. A dedicated team of researchers including bioinformatics scientists, data scientists, medical students, graduate students and summer interns has made contributions to the development of RepurposeDB. The project was under development for last three years, and the first version of the database is now available for public access.
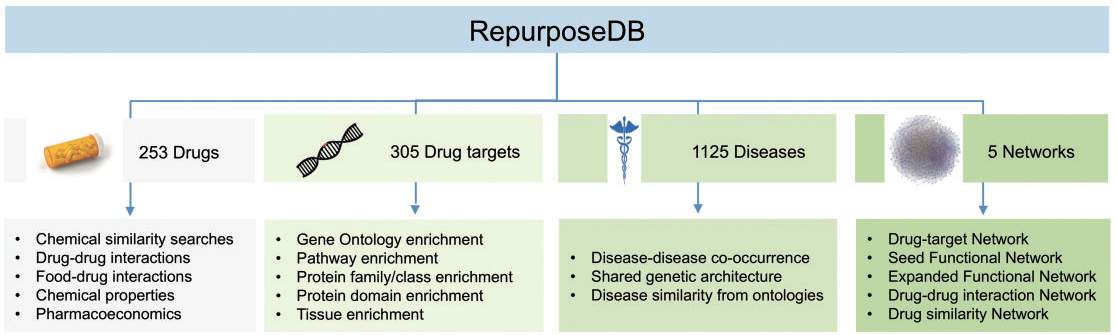
Using the RepurposeDB data, the team has also analyzed and identified drivers of drug repositioning. It is the first time a group is discovering such diverse evidence that could help to design and interpret drug discovery and repositioning studies. For example, by analyzing the commonalities of the small molecule compound library, they discovered chemical moieties, atomic features and side effects that are unique to repurposed compounds. Biological function analyses provide pathways, gene-families, epigenetic factors, protein domains and other biological entities that are common across targets of drug repositioning. The researchers have also compiled data on pair-wise disease similarity and discovered drug repositioning is often successful when disease phenotypes are similar. Also, the team has collected epidemiological data on disease prevalence and identified shared genes across a variety of drug repositioning investigations.
Currently, off-target prescription and drug repositioning studies are reported across scientific literature without a universal standard. As we are entering the Precision Medicine era, it’s important to systematically capture and analyze the data around drug repositioning in a centralized database using an open, community standard. With the user-friendly interface, tools, and crowdsourcing features, RepurposeDB would evolve as an essential resource for the pre-clinical development, medicinal chemistry, systems biology, drug discovery and translational bioinformatics communities.
Link to RepurposeDB: http://repurposedb.dudleylab.org







Leave a Reply